Feature Engineering in in Genomics, Proteomics, and Metabolomics
In the intricate domains of Genomics, Proteomics, and Metabolomics, high-dimensional data presents both a challenge and an opportunity. Identifying biomarkers or class-specific features is not just a computational task but a critical step toward advances like personalized medicine and early disease detection. At Statigen, we've grappled with these challenges across multiple projects, developing advanced techniques to navigate this complex landscape. This blog dives into those techniques, offering you a comprehensive yet nuanced understanding of feature selection in these specialized fields.
The Complexity of High-Dimensional Data
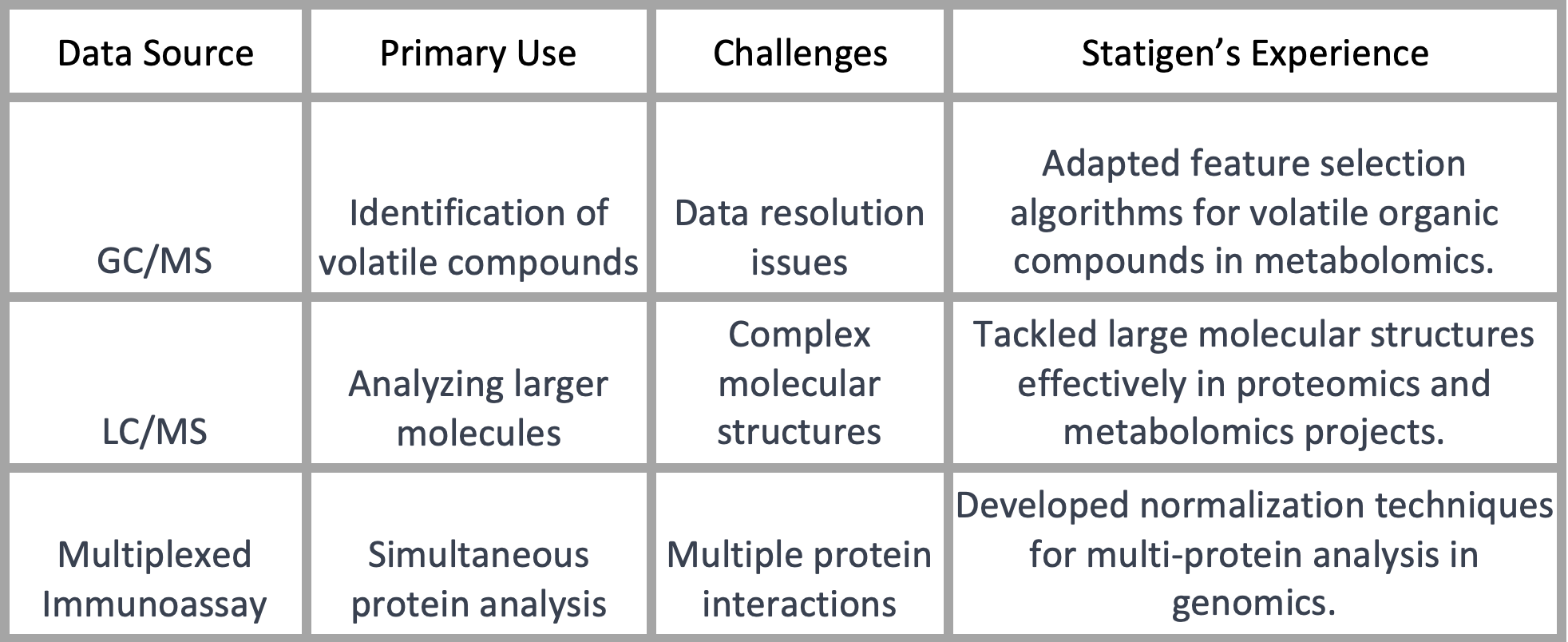
While high-dimensional data is nothing new to data scientists, the types of data encountered in genomics, proteomics, and metabolomics are uniquely challenging. Whether it's data derived from Gas Chromatography-Mass Spectrometry (GC/MS) for metabolomics, Liquid Chromatography-Mass Spectrometry (LC/MS) or multiplexed immunoassays for genomics, each data type comes with its own set of complexities.
At Statigen, we've found that tackling data resolution issues effectively requires a deep understanding of both the hardware and the data's inherent complexities. In projects involving LC/MS data, for example, we had to account for the larger and more complex molecular structures when designing our feature selection algorithms.
Data Preprocessing: A Quick Recap
Any practitioner in AI and ML knows that raw data is seldom ready for immediate analysis, and this holds true in the specialized fields we're discussing. Transforming this raw data into a format amenable to machine learning algorithms is a critical step. At Statigen, we've developed best practices that go beyond standard transformations like Fourier or wavelet transforms. For example, in projects involving genomics, we've had to be particularly cautious about the assumptions made regarding correlations introduced by liquid assays. Our approach ensures that the preprocessing steps enhance, rather than distort, the data's potential for accurate feature selection.
Data Transformation - The Crucial Step
Data transformation is more than just a technical process; it's a pivotal phase that sets the stage for effective feature selection. After the data passes our 'Integrity Filter,' it undergoes a series of transformations. For instance, electrical energy data in the current-voltage domain is translated into frequency space. At Statigen, we've honed our transformation techniques to suit the specific needs of genomic, proteomic, and metabolomic data. Our expertise in understanding the charge transfer process has led us to select basis functions that are most informative for the subsequent steps of feature selection and engineering.
Workflows and Partner Application
Statigen partners with CROs to help them in the analysis of their complex data coming from varied sources like patient records, spectroscopy data and imaging results. This data is deeply rooted in domain-specific knowledge. Given the repetitive nature of clinical data analysis, we incorporated standard recipes into easily accessible workflows. These workflows smoothen the feature selection process and help our partner CROs in drawing clinically relevant insights from data.
The Art and Science of Feature Selection
Feature selection is where the rubber meets the road. It's not merely about choosing variables; it's about making informed decisions that affect the model's performance and interpretability. At Statigen, we've developed a robust methodology that incorporates statistical tests, such as t-tests and several others to rigorously evaluate the significance of each feature. This goes beyond the standard practices and is rooted in our cross-disciplinary expertise. Our methodologies have been particularly effective in reducing the margin of error and enhancing the power of our machine learning models across various projects.
Applications - Where Theory Meets Practice
Understanding the theory behind feature selection is essential, but its real value emerges when applied to various analytical objectives. At Statigen, we've explored multiple facets of these objectives:
For Classification: Our methodology has proven effective in identifying discriminatory features with high statistical power, even when working with synthetic data.
For Quantitation: We've generated highly accurate calibration curves, enhancing the precision of our models in tasks that require measuring the levels of specific biomarkers.
For Anomaly Detection: Identifying anomalies in genomic or proteomic data is crucial for flagging mutations or other rare events significant in a medical or biological context. Our feature selection techniques have been instrumental in these tasks.
For Association: Whether it's Genome-Wide Association Studies (GWAS) in genomics or identifying proteins commonly associated with specific conditions in proteomics, our methodologies have facilitated the uncovering of meaningful relationships in complex data sets.
Conclusion
The complex world of high-dimensional data in genomics, proteomics, and metabolomics demands a multi-faceted approach to feature selection. It's not just about the numbers or the algorithms; it's about connecting computational methods with domain-specific expertise for meaningful results.
At Statigen, we've honed our interdisciplinary skills to navigate these complexities effectively. Our rigorous methodologies don't just address computational challenges but also provide actionable insights, propelling advancements in fields like personalized medicine and early disease detection. Our team is committed to pushing the boundaries of what's possible in genomics, proteomics, and metabolomics.
If you've found this guide insightful, we invite you to explore Statigen's range of services, designed to meet these specialized analytical needs. Stay tuned for more content that delves even deeper into these intricate data landscapes